Analyze everything#
This notebook demonstrates how to run a complete STRESS analysis and produce all relevant output graphs. If you want to download this notebook and execute it locally on your machine, download this file as a ipynb Jupyter notebook file and run it in your local python environment using the download button at the top of this page.
import napari_stress
import napari
import numpy as np
from napari_stress import reconstruction, measurements, TimelapseConverter, utils, stress_backend, plotting
import os
import datetime
from skimage import io
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib as mpl
import yaml
viewer = napari.Viewer(ndisplay=3)
reconstruction_parameters = None
measurement_parameters = None
Load the data#
Replace the following code with the commented out part (and remove the rest) below to load your own data for analysis:
image = napari_stress.get_droplet_4d()[0][0]
image.shape
filename = None
## Replace this code with a command to import your data. Example:
# filename = 'path/to/data.tif'
# image = io.imread(filename)
Data dimensions#
You need to set a few parameters pertaining to your data:
voxel_size_x = 2.078 # microns
voxel_size_y = 2.078 # microns
voxel_size_z = 3.998 # microns
target_voxel_size = 2.078 # microns
time_step = 3 # minutes
viewer.add_image(image, scale=[voxel_size_z, voxel_size_y, voxel_size_x], name='droplet')
<Image layer 'droplet' at 0x19610584bb0>
Analysis parameters#
In case you ran the reconstruction previously interactively from the napari viewer (as explained here) and exported the settings, you can import the settings here, too. To do so, simply uncomment the line below (remove the #) and provide the path to the saved settings file:
# reconstruction_parameters = utils.import_settings(file_name='path/of/reconstruction/settings.yaml')
# measurement_parameters = utils.import_settings(file_name='path/of/measurement/settings.yaml')
If you used a parameter file, you can skip the next step. Otherwise, use this cell to provide the necessary parameters for the reconstruction and the measurement. The parameters are explained here:
If you used the previous cell to import some parameters, skip the next cell or delete it.
reconstruction_parameters = {
'voxelsize': np.asarray([voxel_size_z, voxel_size_y, voxel_size_x]),
'target_voxelsize': target_voxel_size,
'smoothing_sigma': 1,
'n_smoothing_iterations': 15,
'n_points': 256,
'n_tracing_iterations': 3,
'resampling_length': 1,
'fit_type': 'fancy', # can be 'fancy' or 'quick'
'edge_type': 'interior', # can be 'interior' or 'surface'
'trace_length': 20,
'sampling_distance': 0.5,
'interpolation_method': 'cubic', # can be 'linear' 'cubic' or 'nearest'
'outlier_tolerance': 1.5,
'remove_outliers': True,
'return_intermediate_results': True}
measurement_parameters = {
'max_degree': 20, # spherical harmonics degree
'n_quadrature_points': 590, # number of quadrature points to measure on (maximum is 5810)
'gamma': 3.3} # interfacial tension of droplet
alpha = 0.05 # lower and upper boundary in cumulative distribution function which should be used to calculate the stress anisotropy
Hint: If you are working with timelapse data, it is recommended to use parallel computation to speed up the analysis.
parallelize = True
Analysis#
n_frames = image.shape[0]
We run the reconstruction and the stress analysis:
results_reconstruction = reconstruction.reconstruct_droplet(image, **reconstruction_parameters, use_dask=parallelize)
for res in results_reconstruction:
layer = napari.layers.Layer.create(res[0], res[1], res[2])
viewer.add_layer(layer)
Dask client up and running <Client: 'tcp://127.0.0.1:50124' processes=4 threads=4, memory=31.96 GiB> Log: http://127.0.0.1:8787/status
_ = stress_backend.lbdv_info(Max_SPH_Deg=measurement_parameters['max_degree'],
Num_Quad_Pts=measurement_parameters['n_quadrature_points'])
input_data = viewer.layers['Droplet pointcloud (smoothed)'].data
results_stress_analysis = measurements.comprehensive_analysis(input_data, **measurement_parameters,
use_dask=parallelize)
for res in results_stress_analysis:
layer = napari.layers.Layer.create(res[0], res[1], res[2])
viewer.add_layer(layer)
Dask client already running <Client: 'tcp://127.0.0.1:50124' processes=4 threads=4, memory=31.96 GiB> Log: http://127.0.0.1:8787/status
To get an idea about the returned outputs and which is stored in which layer, let’s print them:
for res in results_stress_analysis:
print('-->', res[1]['name'])
if 'metadata' in res[1].keys():
for key in res[1]['metadata'].keys():
print('\t Metadata: ', key)
if 'features' in res[1].keys():
for key in res[1]['features'].keys():
print('\t Features: ', key)
--> Result of fit spherical harmonics (deg = 20)
Metadata: Elipsoid_deviation_contribution_matrix
Metadata: frame
Features: fit_residue
Features: frame
--> Result of expand points on ellipsoid
Features: fit_residue
Features: frame
--> Result of least squares ellipsoid
--> Result of lebedev quadrature on ellipsoid
Metadata: Tissue_stress_tensor_cartesian
Metadata: Tissue_stress_tensor_elliptical
Metadata: Tissue_stress_tensor_elliptical_e1
Metadata: Tissue_stress_tensor_elliptical_e2
Metadata: Tissue_stress_tensor_elliptical_e3
Metadata: stress_ellipsoid_anisotropy_e12
Metadata: stress_ellipsoid_anisotropy_e23
Metadata: stress_ellipsoid_anisotropy_e13
Metadata: angle_ellipsoid_cartesian_e1_x1
Metadata: angle_ellipsoid_cartesian_e1_x2
Metadata: angle_ellipsoid_cartesian_e1_x3
Metadata: stress_tissue_anisotropy
Metadata: frame
Features: mean_curvature
Features: stress_tissue
Features: frame
--> Result of lebedev quadrature (droplet)
Metadata: Gauss_Bonnet_relative_error
Metadata: Gauss_Bonnet_error
Metadata: Gauss_Bonnet_error_radial
Metadata: Gauss_Bonnet_relative_error_radial
Metadata: H0_volume_integral
Metadata: H0_arithmetic
Metadata: H0_surface_integral
Metadata: S2_volume_integral
Metadata: stress_cell_nearest_pair_anisotropy
Metadata: stress_cell_nearest_pair_distance
Metadata: stress_cell_all_pair_anisotropy
Metadata: stress_cell_all_pair_distance
Metadata: frame
Features: mean_curvature
Features: difference_mean_curvature_cartesian_radial_manifold
Features: stress_cell
Features: stress_total
Features: stress_total_radial
Features: stress_cell_extrema
Features: stress_total_extrema
Features: frame
--> Total stress: Geodesics maxima -> nearest minima
--> Total stress: Geodesics minima -> nearest maxima
--> Cell stress: Geodesics maxima -> nearest minima
--> Cell stress: Geodesics minima -> nearest maxima
--> stress_autocorrelations
Metadata: autocorrelations_spatial_total
Metadata: autocorrelations_spatial_cell
Metadata: autocorrelations_spatial_tissue
Metadata: frame
To make handling further down easier, we store all data and metadata in a few simple dataframes
# Compile data
df_over_time, df_nearest_pairs, df_all_pairs, df_autocorrelations = utils.compile_data_from_layers(
results_stress_analysis, n_frames=n_frames, time_step=time_step)
Visualization#
In this section, we will plot some interesting results and save the data to disk. The file location will be at the
%%capture
figures_dict = plotting.create_all_stress_plots(
results_stress_analysis,
time_step=time_step,
n_frames=n_frames
)
mpl.style.use('default')
colormap_time = 'flare'
if filename is not None:
filename_without_ending = os.path.basename(filename).split('.')[0]
save_directory = os.path.join(os.path.dirname(filename), filename_without_ending + '_' + datetime.datetime.now().strftime('%Y-%m-%d_%H-%M-%S'))
else:
save_directory = os.path.join(os.getcwd(), 'results_' + datetime.datetime.now().strftime('%Y-%m-%d_%H-%M-%S'))
if not os.path.exists(save_directory):
os.makedirs(save_directory)
Fit errors#
We first show all the errors that were calculated during the pointcloud refinement:
fit_error_df = pd.DataFrame(results_reconstruction[3][1]['features'].reset_index())
fit_error_df
| index | center | amplitude | slope | background_slope | offset | center_err | amplitude_err | slope_err | background_slope_err | offset_err | distance_to_nearest_neighbor | mean_squared_error | fraction_variance_unexplained | fraction_variance_unexplained_log | idx_of_border | frame | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 20.038063 | 176.410822 | -0.673940 | -1.168413 | 4.609321e-21 | 0.046695 | 2.582717 | 0.012938 | 0.307859 | 0.566758 | 1.099930 | 20.261058 | 0.004435 | -5.418143 | 20.038063 | 0 |
| 1 | 1 | 20.022966 | 163.568899 | -0.638404 | 0.379437 | 5.848198e-20 | 0.047042 | 2.414873 | 0.011207 | 0.275440 | 0.485870 | 1.096523 | 6.302603 | 0.001248 | -6.686067 | 20.022966 | 0 |
| 2 | 2 | 20.025295 | 193.242245 | -0.673258 | -2.377362 | 1.104042e-18 | 0.044560 | 2.653223 | 0.012399 | 0.321607 | 0.599573 | 1.118846 | 33.819470 | 0.007118 | -4.945105 | 20.025295 | 0 |
| 3 | 3 | 20.025143 | 163.229563 | -0.641114 | 0.257542 | 1.242458e-21 | 0.047908 | 2.453894 | 0.011555 | 0.280890 | 0.497471 | 1.031137 | 10.036452 | 0.002015 | -6.207211 | 20.025143 | 0 |
| 4 | 4 | 20.003009 | 176.166124 | -0.670780 | -0.900823 | 3.622026e-20 | 0.048619 | 2.688943 | 0.013216 | 0.320005 | 0.577818 | 1.076207 | 10.774460 | 0.002298 | -6.075505 | 20.003009 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15364 | 725 | 19.998927 | 197.392632 | -0.627527 | -3.770612 | 3.676951e-01 | 0.018876 | 0.962403 | 0.004785 | 0.103900 | 0.237937 | 1.031561 | 17.373360 | 0.003571 | -5.634947 | 19.998927 | 20 |
| 15365 | 726 | 20.040552 | 171.129640 | -0.677093 | -1.781867 | 3.194370e-14 | 0.029979 | 1.459065 | 0.009372 | 0.152425 | 0.390491 | 1.131636 | 18.176025 | 0.003844 | -5.561195 | 20.040552 | 20 |
| 15366 | 727 | 19.995654 | 179.672688 | -0.667468 | -2.447113 | 6.488004e-22 | 0.025294 | 1.271959 | 0.007641 | 0.134078 | 0.340907 | 1.085043 | 15.459548 | 0.003339 | -5.702091 | 19.995654 | 20 |
| 15367 | 728 | 19.992411 | 192.269531 | -0.652320 | -3.107330 | 1.823652e-16 | 0.023571 | 1.247022 | 0.006770 | 0.131471 | 0.335464 | 1.074869 | 19.429097 | 0.003838 | -5.562913 | 19.992411 | 20 |
| 15368 | 729 | 20.031783 | 169.546593 | -0.675959 | -1.751943 | 1.263994e-20 | 0.030450 | 1.467567 | 0.009474 | 0.153350 | 0.391583 | NaN | 19.560836 | 0.004306 | -5.447669 | 20.031783 | 20 |
15369 rows × 17 columns
fig, axes = plt.subplots(ncols=4, nrows=len(fit_error_df.columns)//4+1, figsize=(20, 10), sharey=True)
axes = axes.flatten()
for idx, column in enumerate(fit_error_df.columns):
ax = axes[idx]
sns.histplot(data=fit_error_df, x=column, ax=ax, bins=100)
ax.set_xlabel(column, fontsize=16)
ax.set_ylabel('Counts [#]', fontsize=16)
if save_directory is not None:
fig.savefig(os.path.join(save_directory, 'fit_error_reconstruction.png'), dpi=300)
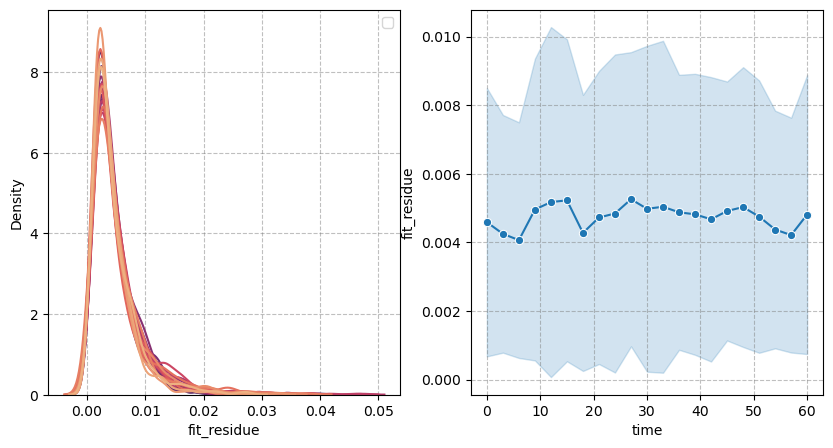
Spherical harmonics#
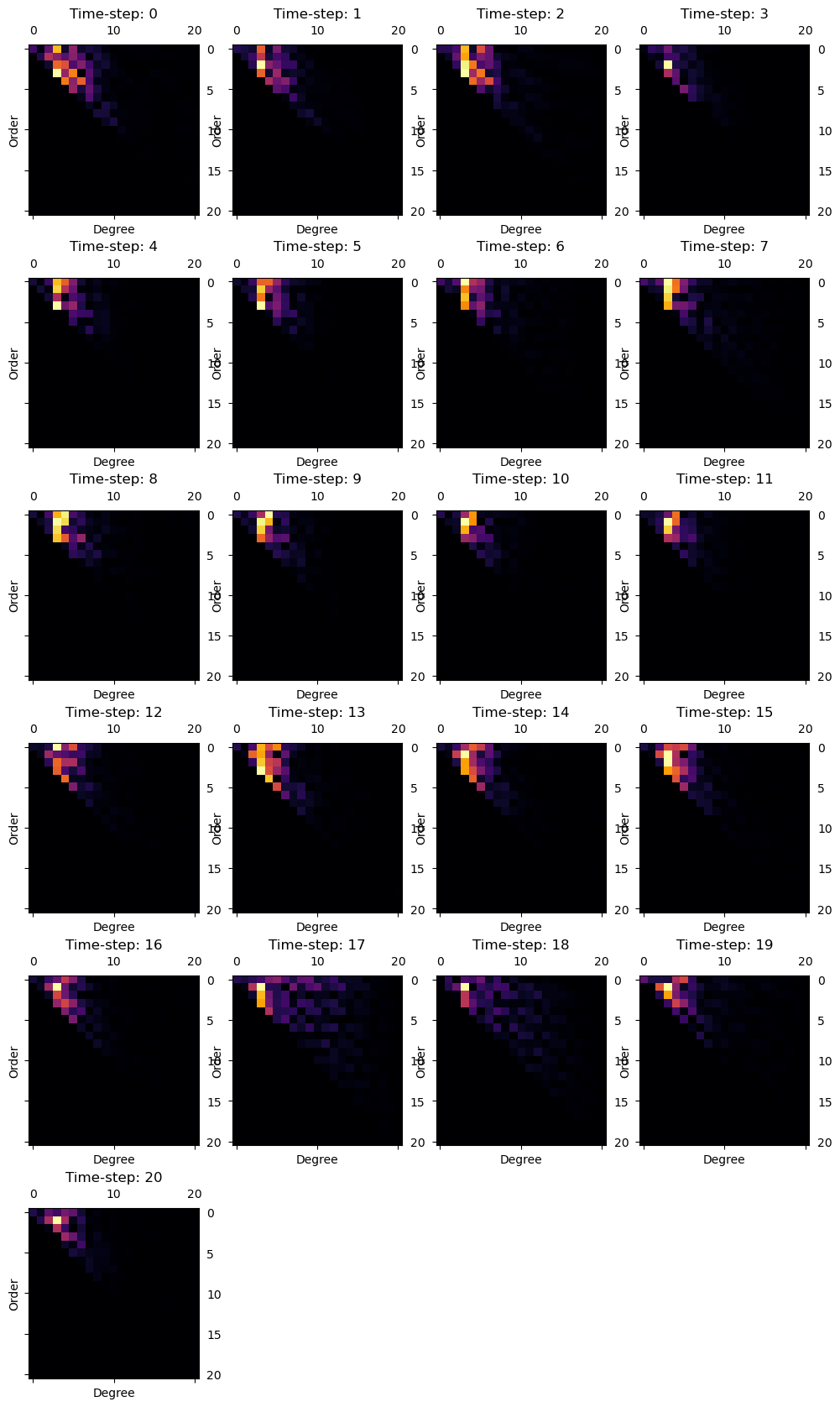
Fit residue#
We now show the errors made when approximating the reconstructed pointcloud with the spherical harmonics:
figure = figures_dict['Figure_reside']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
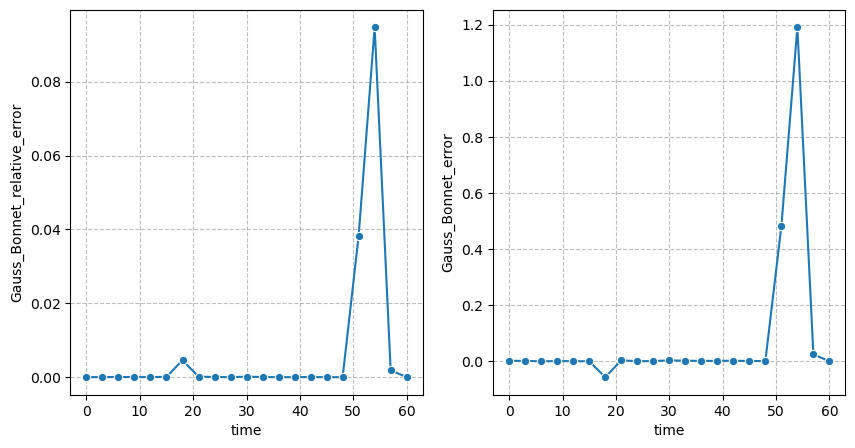
Fit quality#
We can quantify the quality of the extracted pointcloud by using the absolute and relative Gauss-Bonnet errors:
figure = figures_dict['fig_GaussBonnet_error']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
Curvature#
We next show mean curvature histograms and averages over time:
figure = figures_dict['fig_mean_curvature']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure'].axes[0].set_xlabel("Mean curvature [$mm^{-1}$]", fontsize=16)
figure['figure'].axes[1].set_ylabel("Mean curvature [$mm^{-1}$]", fontsize=16)
figure['figure'].axes[1].set_xlabel("Time [min]", fontsize=16)
figure['figure']
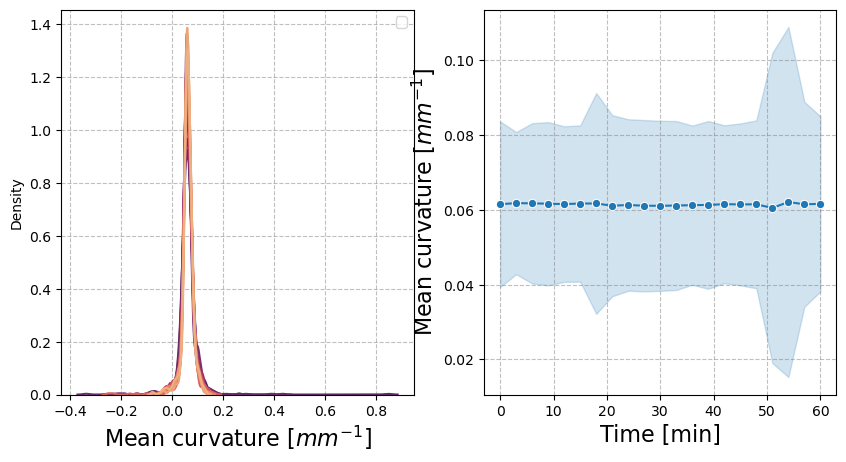
figure = figures_dict['fig_total_stress']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
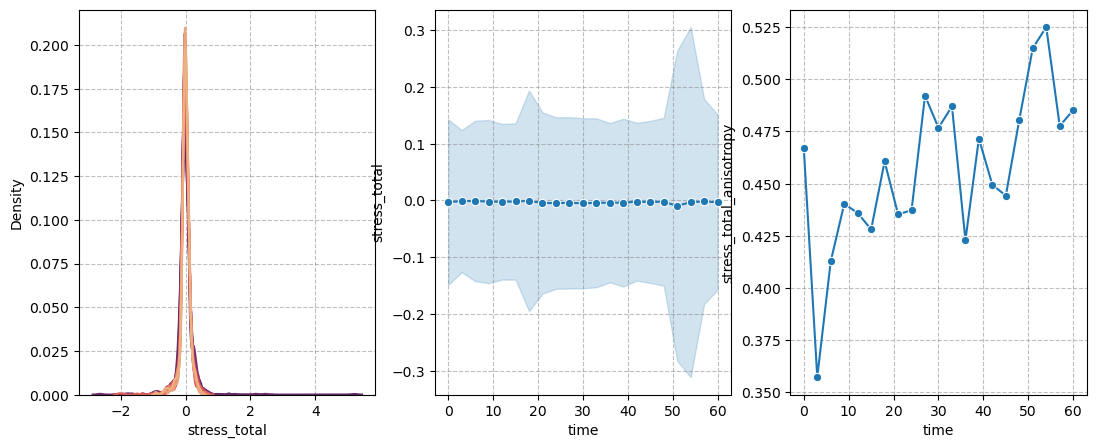
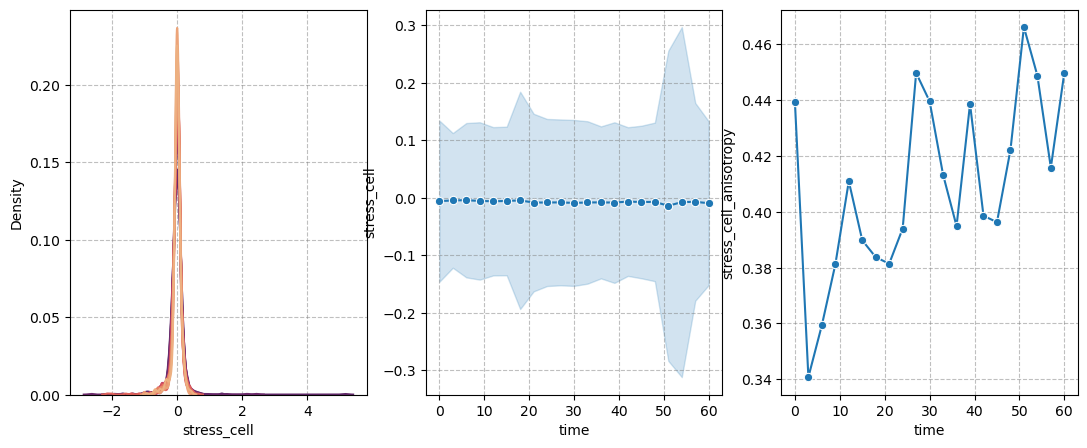
figure = figures_dict['fig_cell_stress']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
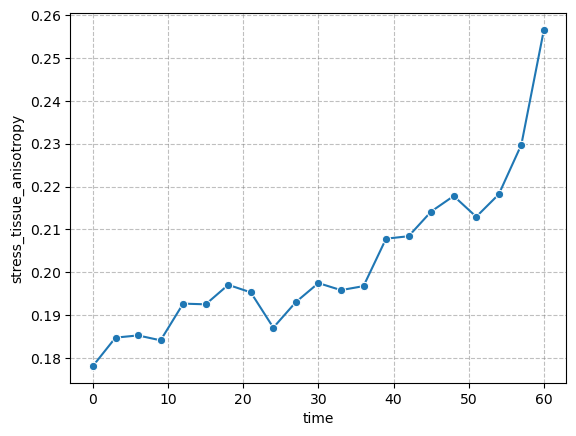
Tissue-scale stresses#
figure = figures_dict['fig_tissue_stress']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
Stress along axes#
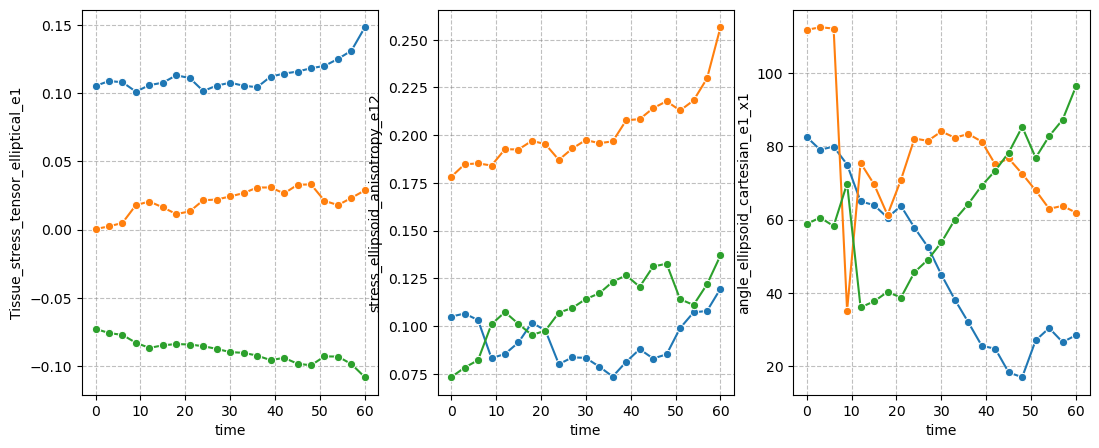
figure = figures_dict['fig_stress_tensor']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
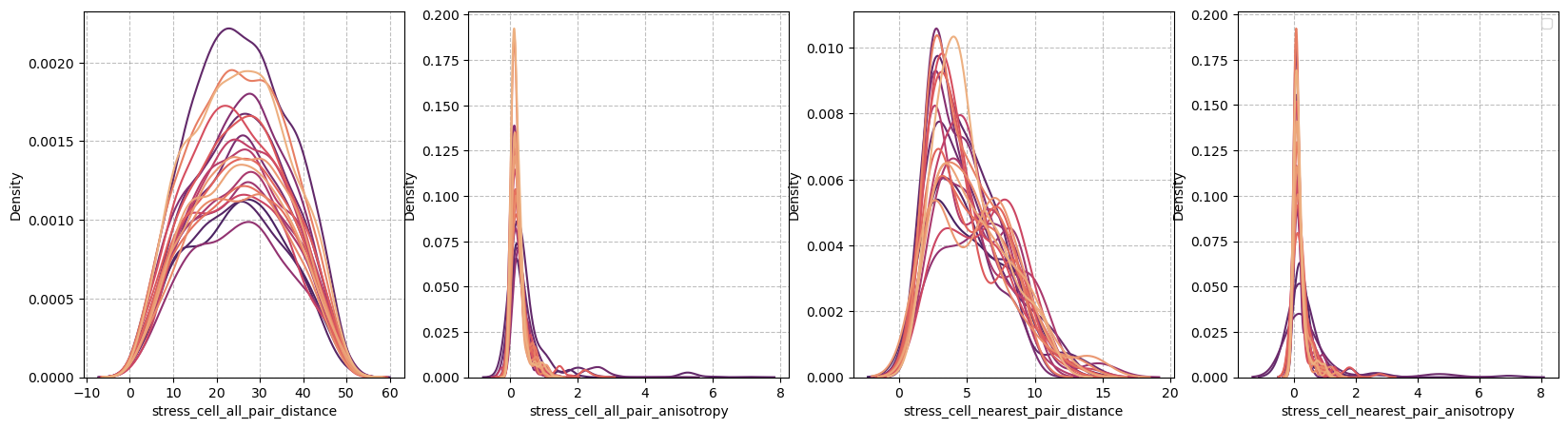
Extrema analysis#
figure = figures_dict['fig_all_pairs']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
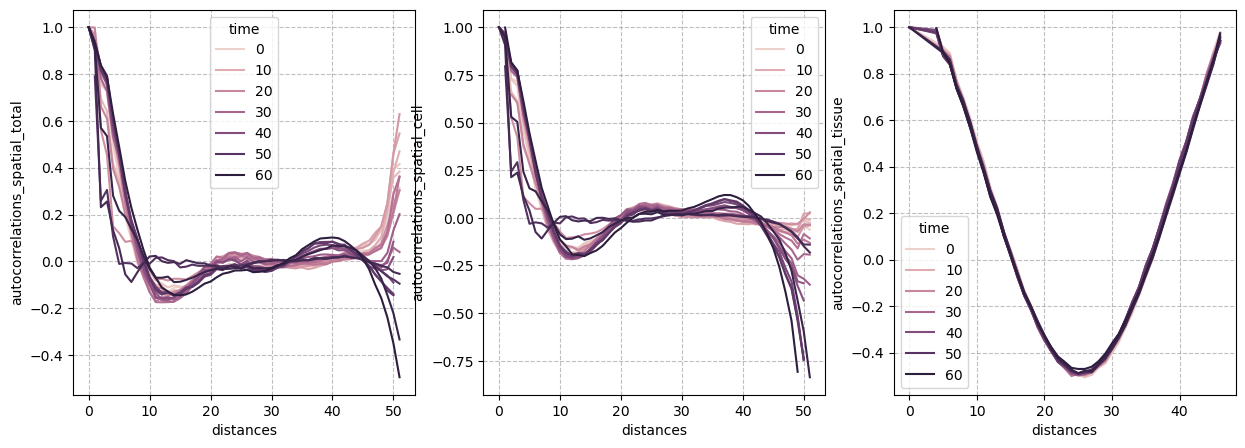
Autocorrelations: Spatial#
figure = figures_dict['fig_spatial_autocorrelation']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
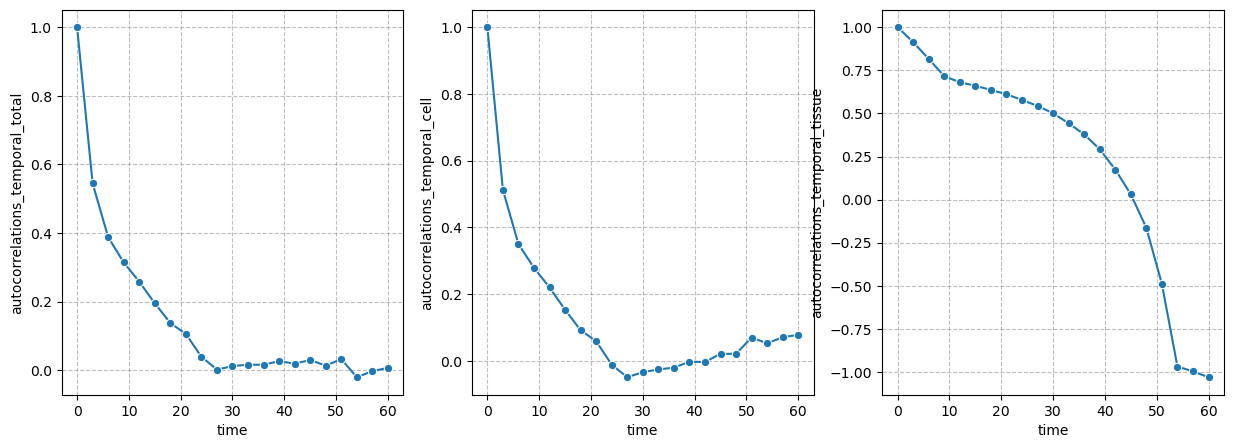
Autocorrelations: Temporal#
figure = figures_dict['fig_temporal_autocorrelation']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
Ellipsoid deviation#
figure = figures_dict['fig_ellipsoid_contribution']
if save_directory is not None:
figure['figure'].savefig(os.path.join(save_directory, figure['path']), dpi=300)
figure['figure']
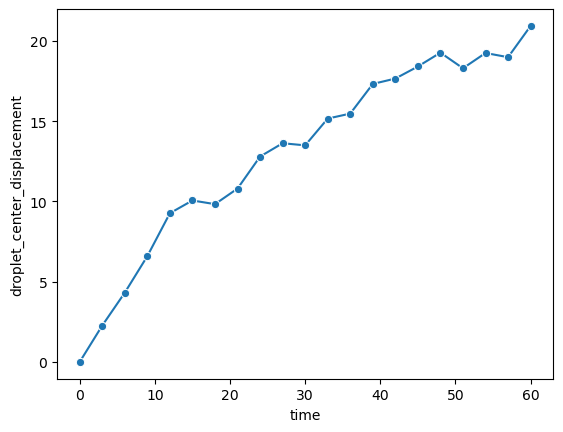
Droplet movement#
This analyzes how much the center of the droplet moves over time.
Converter = TimelapseConverter()
list_of_points = Converter.data_to_list_of_data(results_reconstruction[3][0], layertype=napari.types.PointsData)
center = [np.mean(points, axis=0) for points in list_of_points]
center_displacement = np.asarray([np.linalg.norm(center[t] - center[0]) for t in range(n_frames)])
df_over_time['droplet_center_displacement'] = center_displacement * target_voxel_size
sns.lineplot(data=df_over_time, x='time', y='droplet_center_displacement', marker='o')
<Axes: xlabel='time', ylabel='droplet_center_displacement'>
Export data#
We first agregate the data from the spatial autocorrelations in a separate dataframe. This dataframe has a column for autocorrelations of total, cell and tissue-scale stresses.
df_to_export = pd.DataFrame()
for col in df_over_time.columns:
if isinstance(df_over_time[col].iloc[0], np.ndarray):
continue
if np.stack(df_over_time[col].to_numpy()).shape == (n_frames,):
df_to_export[col] = df_over_time[col].to_numpy()
df_to_export.to_csv(os.path.join(save_directory, 'results_over_time.csv'), index=False)
We also export the used settings for the analysis into a .yml file:
utils.export_settings(reconstruction_parameters, file_name=os.path.join(save_directory, 'reconstruction_settings.yaml'))
utils.export_settings(measurement_parameters, file_name=os.path.join(save_directory, 'measurement_settings.yaml'))